| A | B |
|---|
| In both prokaryotes and eukaryotes, gene expression is most often regulated at the stage of _______. | transcription,  |
| The condensed form of chromatin is called a(n) _______. | chromosome (The karyotype below shows the 46 human chromosomes visible during mitosis),  |
| The condensed form of ______ is called a chromosome. | chromatin |
| DNA wraps around ________ to form chromatin. | histone proteins, 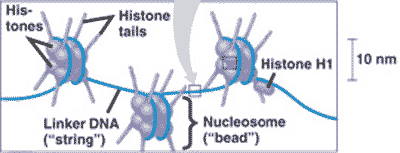 |
| During development of multicellular organisms, cells undergo a process of becoming specialized in form and function in a process called ________. | cell differentiation (this involves activating certain genes and permanently silencing others, such as genes for liver proteins in a pancreas cell) |
| Differences between cell types are due not to different genes being present in the cell but to ______. | differential gene expression |
| What affect does the acetylation of histone tails have on the ability of DNA to be transcribed? | makes it easier (because the acetyl group -COCH3 neutralizes the positive charge of the histone protein, loosening the grip of the histone on the negatively charged DNA. It also loosens their attraction to neighboring nucleosomes), 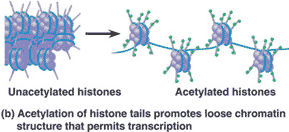 |
| What affect does the methylation of histone tails have on the ability of DNA to be transcribed? | makes it harder |
| DNA that is ______ is generally inactive. | methylated |
| Once methylated, genes usually ______ through successive cell divisions. | stay that way (this can permanently silence genes that arise from the methylated parent cell) |
| Genomic imprinting, where genes are expressed depending on the sex of the parent, is caused by ______ of maternal or paternal alleles early in the development process, affectively shutting the alleles down even in daughter cells that are formed from then on. | methylation |
| Inheritance of traits transmitted by mechanisms not directly involving the nucleotide sequence is called ____ inheritance. Methylation, which can silence genes even in successive cell divisions, is an example. | epigenetic inheritance |
| The cluster of proteins that assembles on the promoter sequence at the upstream end of a gene is called the _____. | transcription initiation complex,  |
| Segments of non-coding DNA that help regulate transcription by binding certain proteins are called ______. | control elements,  |
| To initiate transcription, eukaryotic RNA polymerase requires the assistance of proteins called ______. | transcription factors,  |
| Transcription factors that are essential for the transcription of ALL protein-coding genes are called ______ transcription factors. In order to get high levels of transcription of a particular gene, additional _________ transcription factors are required. | general, specific (the activators in this diagram are a type of specific transcription factor),  |
| Distal control elements collectively are also called _____. | enhancers,  |
| Enhancers are made up of multiple _______. | distal control elements,  |
| Enhancers (a.k.a. distal control elements) interact with two types of specific transcription factors, ______ and ______. | activators and repressors,  |
| In eukaryotes, the precise control of transcription depends largely on the binding of proteins called _______ to DNA segments called control elements. | activators,  |
| Name four mechanisms of post-transcriptional regulation of gene expression. | 1) RNA processing 2) mRNA degradation 3) Initiation of translation 4) Protein processing and degradation |
| One example of regulation at the RNA processing level is _______, in which different mRNA molecules are produced from the same primary transcript, depending on which RNA segments are used as exons and which are used as introns. | alternative RNA splicing, 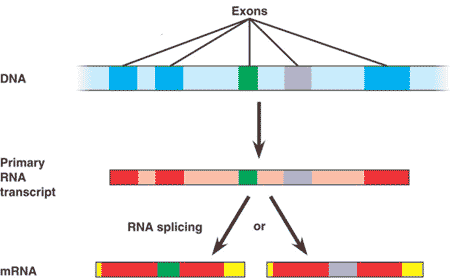 |
| What affects the length of time that mRNA survives in the cytoplasm? | nucleotide sequences in the 3' UTR |
| What are two effects that microRNA's (miRNA) can have on mRNA? | 1) It can bind to mRNA, blocking its translation 2) It can bind to mRNA and degrades it (both process require the binding of a protein to the miRNA's, 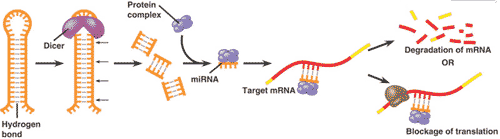 |
| The initiation of translation of selected mRNA's can be blocked by regulatory proteins that bind to specific sequences or structures within the _______ of mRNA. | 5' UTR (untranslated region), 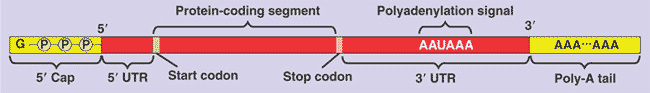 |
| To mark a particular protein for destruction, the cell commonly attaches molecules of a small protein called ______ to the protein. Giant protein complexes called ______ then recognize the tagged protein and degrades it. | ubiquitin, proteasomes,  |
| General transcription factors function in assembling the _______ at the promoters for all genes while specific transcription factors bind to control elements and either increase (if they are _______) or decrease (if they are _______) transcription of that gene. | transcription initiation complex; activators, repressors,  |
| The genes that normally regulate cell growth and division include ______, their _______, and ________ molecules. Mutations that alter any of these genes in somatic cells can lead to _____. | growth factors, their receptors, signaling pathway molecules, cancer |
| The Epstein-Barr virus, Papilloma virus and HTLV-1 virus have all been identified as ____ viruses that cause ______. | tumor viruses, cancer |
| Cancer causing genes are called _____. | oncogenes |
| ______ code for proteins that stimulate normal cell growth and division, but can be turned into a cancer-causing gene by mutations. | proto-oncogenes |
| ______ genes encode for proteins that help prevent uncontrolled cell growth. | Tumor-suppressor genes |
| The first sign of cancer may be a small benign growth called a(n) ______. | polyp, 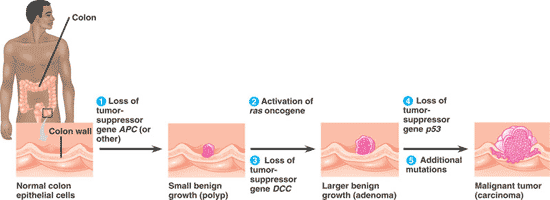 |
| A tumor larger than a polyp, but still benign is called a(n) _____. | adenoma (a.k.a. benign tumor),  |
| A tumor large enough to impair function of one or several organs is called a _____ or ______. | carcinoma, malignant tumor,  |
| Malignant tumor cells tend to produce an enzyme called ____ which keeps its telomeres from shortening and gives its cells the ability to divide indefinitely. | telomerase |
| An individual ______ an oncogene or mutant allele of a tumor-suppressor gene is one step closer to accumulating the necessary mutations for cancer to develop by the _____ model of cancer development. | inheriting, multistep,  |
The diagram below shows the movement of a(n) _____ within the genome., 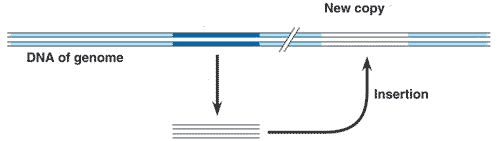 | transposon,  |
The diagram below shows the movement of a(n) _____ within the genome.,  | retrotransposon,  |
What is "A" pointing to in the picture below?, 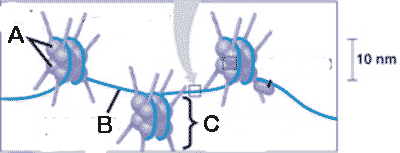 | histone proteins,  |
What is "B" pointing to in the picture below?,  | linker DNA,  |
What is "C" pointing to in the picture below?,  | a nucleosome,  |
| The more open, unraveled form of eukaryotic chromatin that is available for transcription. | euchromatin |
| A gene found in viri or as part of the normal genome that is involved in triggering cancerous characteristics. | oncogene |
| A small, single-stranded RNA molecule that binds to a complementary sequence in mRNA molecules and directs associated proteins to degrade or prevent translation of the target mRNA. | miRNA (microRNA) |
| A giant protein complex that recognizes and destroys proteins tagged for elimination by the small protein ubiquitin. | A proteasome,  |
| Nucleotide sequences, usually noncoding, that are present in many copies in a eukaryotic genome. The repeating units may be short and arranged tandemly (in series) or long and dispersed in the genome. | repetitive DNA |
| The guardian angel of the genome, this gene is expressed when a cell's DNA is damaged. Its product is a transcription factor that transcribes proteins whose job it is to keep mutated cells from dividing or help repair damaged DNA. | p53 gene, 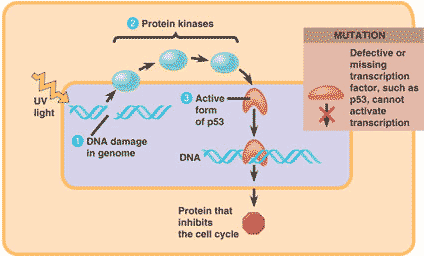 |
| A gene that codes for a type of G protein that relays a growth signal from a growth factor receptor on the plasma membrane to a cascade of protein kinases that ultimately results in the stimulation of the cell cycle. Point mutations in this gene can lead to a hyperactive version of the protein that can lead to excessive cell division and cancer. | Ras gene,  |
| A segment of noncoding DNA that helps regulate transcription of a gene by binding proteins called transcription factors. | control element,  |
| Expression of different sets of genes by cells with the same genome is called _____. | differential gene expression |
| Nontranscribed eukaryotic chromatin that is so highly compacted that it is visible with a light microscope during interphase. | heterochromatin |
| A protein that suppresses the transcription of a gene is called a(n) _____. | repressor |
| A normal cellular gene corresponding to an oncogene; a gene with the potential to cause cancer, but that requires some alteration to become an oncogene. | proto-oncogene |
| A DNA segment very similar to a real gene but which does not yield a functional product; a gene that has become inactivated in a particular species because of a mutation. | pseudogene ("pseudo" means "false") |
| A collection of genes with similar or identical sequences, presumably of common origin. | multigene family |
| The phenomenon in which expression of an allele in offspring depends on whether the allele is inherited from the male or the female parent. | genomic imprinting (this is often accomplished by methylation of an allele early in development) |
| A technique to silence the expression of selected genes in non-mammalian organisms. The method uses synthetic double-stranded RNA molecules matching the sequence of a particular gene to trigger the breakdown of the gene's messenger RNA. | RNA interference |
| The structural and functional divergence of cells as they become specialized during a multicellular organism's development; depending on the control of gene expression. | cell differentiation |
| A gene whose protein products inhibit cell division, thereby preventing uncontrolled cell growth (cancer). | tumor-suppressor gene |
| What do you call the molecule that results from the injection of small double stranded RNA into non-mammalian cells by scientists in order to silence genes with the same sequence? | siRNA's (small interfering RNA's). <siRNA's seem to be formed the same way that microRNAs (miRNAs) are formed naturally within a cell. Both have the same effect of silencing genes> |