| A | B |
|---|
| The enzyme that catalyzes the elongation of new DNA at a replication fork by the addition of nucleotides to the existing chain is called _____. | DNA polymerase (the orange molecules in the picture below) p314,  |
| The enzyme that joins RNA nucleotides to make the primer is called ____. | primase (letter B in the picture below) p316,  |
| The enzyme that untwists and unzips the double helix of DNA at the replication forks is called ____. | helicase (letter A in the picture below) p314,  |
| A protein that functions in DNA replication, helping to relieve strain in the double helix ahead of the replication fork is called ____. | topoisomerase p314 |
| An enzyme that hydrolyzes DNA and RNA into their component nucleotides, and is used to cut out damaged DNA during nucleotide excision repair, is called _______. | a nuclease p318 |
| _____ is a linking enzyme essential for DNA replication. It catalyzes the covalent bonding of the 3' end of a new DNA fragment to the 5' end of a growing chain. Specifically, it bonds the sugar from one nucleotide to the phosphate of another nucleotide. | DNA ligase p316,  |
| The change in genotype and phenotype due to the assimilation of external DNA by a cell is called ____. | transformation (This use of the word transformation should not be confused with the conversion of a normal animal cell to a cancerous one, discussed in section 12.3) p306 |
| A short segment of DNA synthesized on a template strand during DNA replication which is joined to other short segments to form the lagging strand is called a(n) _____. | Okazaki fragment p315,  |
| The cellular process that uses special enzymes to fix incorrectly paired nucleotides that were not corrected by DNA polymerase is called ____. | mismatch repair p317 |
| During DNA replication, _________, are molecules that line up along the unpaired DNA strands, holding them apart while the DNA strands serve as templates for the synthesis of complementary strands of DNA. | single-strand binding proteins (letter D in the picture below) p314,  |
| The ______ is the site where the replication of a DNA molecule begins. | origin of replication p313 |
| The process of copying a cell's DNA is called _______. | DNA replication p311,  |
| The shape of a DNA molecule is referred to as a(n) _____. | double helix p309,  |
| The enzyme that catalyzes the lengthening of telomeres in germ cells (and many cancer cells) is called _____. | telomerase (This enzyme keeps the telomeres full length in germ cells so that the full length chromosomes can be passed on to offspring. Somatic cells get shorter and shorter at each cell division and eventually, the shortening of the DNA effects not only the telomeres, which act as buffer, but actual genes, thereby limiting the number of cell divisions that somatic cells can undergo during their lifetime) p318 |
| The new continuous complementary DNA strand synthesized along the template strand in the mandatory 5' to 3' direction. | leading strand pp315-316,  |
| The process of removing and then correctly replacing a damaged segment of DNA using the undamaged strand as a guide is called _____. | nucleotide excision repair p318, 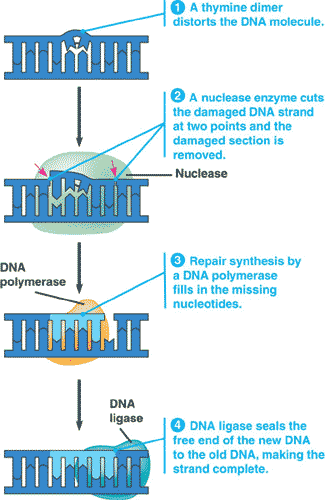 |
| A virus that infects bacteria is called a(n) _____. | bacteriophage p306,  |
| The protective structure at each end of a eukaryotic chromosome that consists of repetitive extra DNA. | telomere p318 |
| The y-shaped region on a replicating DNA molecule where new strands are growing is called the ______. | replication fork p314,  |
| The discontinuously synthesized DNA strand that elongates in a direction away from the replication fork is called the ______. | lagging strand p315,  |
The picture below shows a(n) _____.,  | bacteriophage (this virus infects bacteria only) p306,  |
What type of bonds (represented by the pink dotted lines) hold the bases of complementary DNA together?,  | hydrogen bonds p310,  |
Which nucleotides below are purines and which are pyrimidines?,  | Thymine and Cytosine are the pyrimidines and Adenine and Guanine are the purines (You might be able to remember that pyrimidines are the ones with a single ring structure because it would be easier to measure the "perimeter" of a single-ring nitrogenous base. Get it? Perimeter, pyrimidine?) p310,  |
| According to _____, there is the same percentage of adenine and thymine in the genome of a given species and the same percentage of guanine and cytosine. | Chargaff's rules p308 |
Which process is shown in the picture below?,  | nucleotide excision repair p318,  |
The letter A is pointing to the ______.,  | leading strand pp315-316,  |
The letter B is pointing to the ______.,  | lagging strand pp315-316,  |
The letter C is pointing to the ______.,  | Okazaki fragments p315,  |
The letter D is pointing to ______.,  | DNA polymerase p315,  |
The letter E is pointing to the ______.,  | replication fork pp313-316,  |
The letter F is pointing to ______.,  | DNA ligase (notice that it is catalyzing the bonding of the sugar end of one Okazaki fragment to the phosphate end of the other Okazaki fragment) p316,  |
What is the last name of the scientist(s) who performed the experiment shown below?, 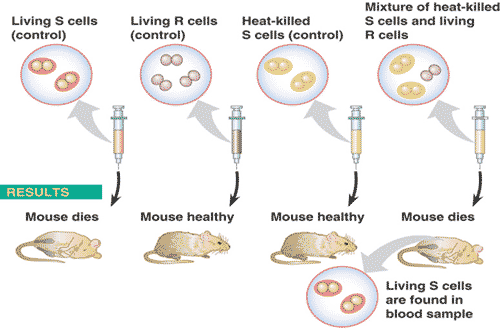 | Griffith (be able to explain this experiment in detail) p306,  |
What is the last name of the scientist(s) who performed the experiment shown below?,  | Hershey and Chase (be able to explain this experiment in detail) p307,  |
The letter A is pointing to ______.,  | helicase (this enzyme unwinds and unzips the DNA molecule) p314,  |
The letter B is pointing to ______.,  | primase (This enzyme lays down the necessary primer nucleotides to get the process of DNA replication going. Only one primer is needed on the leading strand while the lagging strand needs a primer for each Okazaki fragment) pp315-316,  |
The letter C is pointing to ______.,  | a primer (This is a short segment of either DNA or RNA that is needed to initiate DNA synthesis because DNA polymerase can only add nucleotides to existing new strands. In humans, the primer is made of RNA nucleotides which will need to be replaced by DNA nucleotides to complete the strand) pp315-316,  |
The letter D is pointing to ______.,  | Single-strand binding proteins (These are proteins that line up along the unpaired DNA strands, holding them apart while the DNA strands serve as templates for the synthesis of complementary strands of DNA.) pp314,  |
| The nitrogenous base adenine always bonds to ______ in a DNA molecule. | thymine p310 |
| The nitrogenous base cytosine always bonds to ______ in a DNA molecule. | guanine p310 |
| What is the last name of the scientist(s) who used DNA destroying enzymes, RNA destroying enzymes, and protein destroying enzymes to determine that DNA was responsible for the transformation seen in Frederick Griffiths experiment (the one where the mice die). | Avery (Be able to explain Oswald Avery's experiment in detail) p306 |
| Alfred Hershey and Martha Chase used radioactive ______ to label the proteins in the T2 bacteriophage. | sulfur (They used sulfur because it is found in some amino acids that make up protein, but is never found in the nucleotides that make up DNA) pp307&308,  |
| Alfred Hershey and Martha Chase used radioactive ______ to label the DNA in the T2 bacteriophage. | phosphorus (They used phosphorus because it is found in the nucleotides that make up DNA, but is never found in the amino acids that make up proteins) pp307&308,  |
| Which carbon on the pentose sugar of DNA binds to the nitrogenous base? | 1' p308,  |
| Which carbon on the pentose sugar of DNA binds to the phosphate group that completes the nucleotide? | 5' pp308&309,  |
| Which carbon on the pentose sugar of DNA binds to the phosphate group of the next nucleotide in the strand? | 3' (remember that strands are built in the 5' --> 3' direction),  |
What is this a picture of and what is the name of the technique used to produce it?,  | DNA, X-ray crystallography p309,  |
| What was the name(s) of the scientist who first published a paper describing the correct structure of DNA in 1953? | Watson and Crick (They, along with Maurice Wilkins, were awarded the Nobel Prize for their work on determining the structure of DNA. The prize was awarded in 1962. Rosalind Franklin had passed away in 1958 and was therefore ineligible for the prize. Many people believe that Rosalind Franklin did not get the credit she deserved while Watson and Crick got too much.) pp305&310 |
| What was the name of the scientist(s) who produced images of DNA using X-ray crystallography that helped Watson and Crick decipher the structure of DNA? | Rosalind Franklin (Below is an image of DNA that she produced. Watson and Crick, along with Franklin's partner, Maurice Wilkins, were awarded the Nobel Prize for their work on determining the structure of DNA. The prize was awarded in 1962. Rosalind Franklin had passed away in 1958 and was therefore ineligible for the prize. Many people believe that Rosalind Franklin did not get the credit she deserved while Watson and Crick got too much.) p309,  |
Which model of DNA replication is correct and what is the model called?, 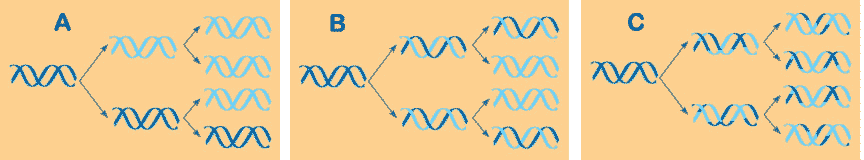 | B) semiconservative model (was worked out by Meselson and Stahl using radioactive nitrogen markers to trace the path of the parent DNA strands) pp311&312,  |
| Which two nucleotides have nitrogenous bases that are classified as pyrimidines? | thymine and cytosine p310, 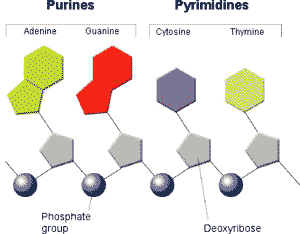 |
| Which two nucleotides have nitrogenous bases that are classified as purines? | adenine and guanine p310,  |
| Which two complementary pairs of nitrogenous bases bond with 2 hydrogen bonds? | thymine and adenine (You might be able to better remember this by the phrase, "A T for two, GC three") p310, 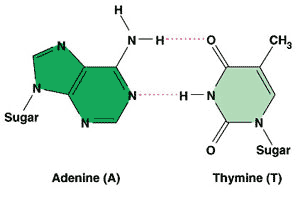 |
| Which two complementary pairs of nitrogenous bases bond with 3 hydrogen bonds? | guanine and cytosine (You might be able to better remember this by the phrase, "A T for two, GC three") p310, 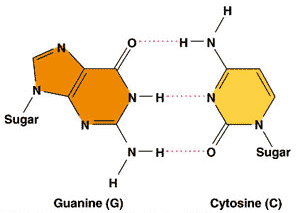 |
| A permanent change in a DNA sequence is called a(n) ______. | mutation p318 |
| DNA wraps around ________ to form chromatin. | histone proteins p320, 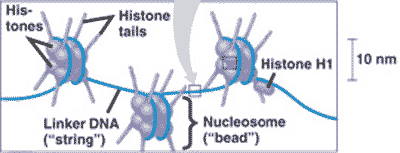 |
What is "A" pointing to in the picture below?, 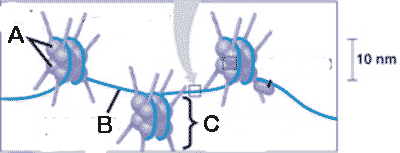 | histone proteins p320,  |
What is "B" pointing to in the picture below?,  | linker DNA p320,  |
What is "C" pointing to in the picture below?,  | a nucleosome p320,  |
| The more open, unraveled form of eukaryotic chromatin that is available for transcription. | euchromatin p322 |
| Nontranscribed eukaryotic chromatin that is so highly compacted that it is visible with a light microscope during interphase. | heterochromatin p322 |